Recently, though unremitting efforts, Dajian ZHANG's research group from SDAU achieved breakthroughs in the field of soybean genome research. The team has analyzed the evolution process of Glycine genome, efficiently and accurately excavated the structural variation of the genus Glycine, broadened the genetic resources available for molecular breeding of cultivated soybean, and provided important theoretical support for the dissection the basis of soybean genetic variation, particularly for mining gene regulated domestication related traits r. On March 15, Beijing time, Nature Plants, a well-known journal in the field of international botany, published this important research result online.

Screenshot of the thesis
Soybean is an important crop, providing both oil and protein, has an important role in the development of the national economy. Cultivated soybeans originated in China, was domesticated by the long-term targeted selection and breeding from ancestral wild soybeans, but in the process of domestication and improvement, only a small amount of genetic resources were manually selected, which results in a serious genetic bottleneck effect and greatly limited the yield and quality improvement of cultivated soybeans. In recent years, China's soybean production has stagnated, imports have continued to grow, and more than 80% of the soybean production depends on foreign imports, which seriously threatens our food security. Thus, the germplasm improvement and breeding of soybean is imminent.
Wild soybean and cultivated soybean
Soybean is a member of Leguminosae family, belongs to Glycine genus, which is divided into two subgenuses, Glycine and Soja. The subgenus Soja is divided into annual wild soybeans (G. soja) and annually cultivated soybeans (G. max). Wild soybeans are an important genetic resource for traits improving of cultivated soybeans. Currently, reference genomes of representative annual soybean germplasm have been reported in the Soja subgenus, and high-quality pan-genomes have been constructed based on graphic structure, which provides an extremely important resource and platform for in-depth study of soybean functional genomics. However, with the rapid development of soybean breeding, soybean germplasm resources are relatively scarce. Perennial wild soybeans have abundant genetic diversity, and excellent biotic/abiotic resistance. The genetic diversity serves as valuable resources for the mining and breeding of important agronomic traits of cultivated soybean. However, due to the large, repetitive sequences rich and high heterozygous characteristics of the perennial wild soybean genomes, construction of high-quality chromosomal-level reference genomes have been successful.

Perennial wild soybean
In the reported study, the researchers selected five representative Glycine species (representing A, B, C, D, F subgenome types in the Glycine subgenus, respectively) and a naturally formed tetraploid (AADD) perennial soybean for whole genome sequencing. By combination of sequencing technologies such as second-generation, third-generation and hi-C. High-quality reference genomes at the chromosomal level were assembled for the six species respectively, and a pan-genome of genus Glycine was constructed for the first time. 109,827 non-redundant gene loci were identified in the perennial soybeans and 70% of them were found to be lost in the subgenus Soja, severing as valuable genetic resources for soybean breeding.

Wild soybean seeds
This is the first construction of the Glycine subgenus genomes and an important breakthrough in the research of perennial wild crops as genetic resources, filled the gaps in the genome of Glycine subgenus, and provides valuable genetic resources for the breeding of high-yield and high-quality varieties of soybean.
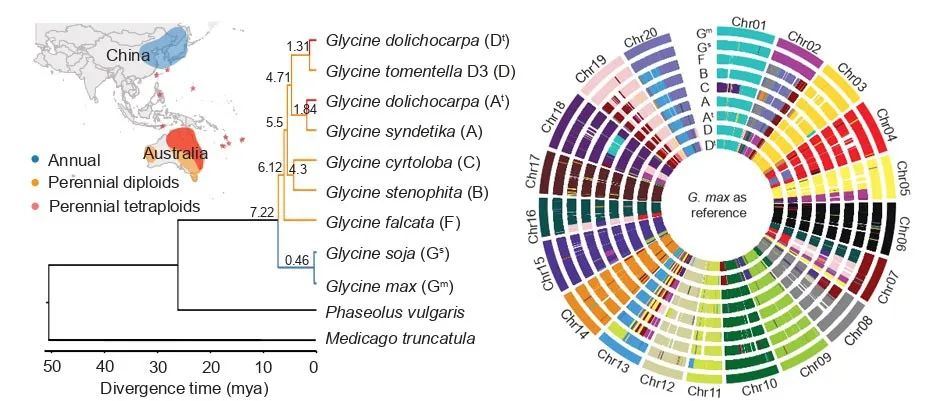
Analysis of the perennial wild soybean genome
Prof. Dajian ZHANG, the corresponding author of the paper, said that in recent years, with the continuous development and progress of molecular biology technology, more and more evidence shows that the structural variation of large fragments is widely present in the crop genome and affects many important agronomic traits. In this study, 183 large-scale genomic structural variations were identified by establishing a collinear relationship between the genomes of the two subgenus, which affected important phenotypic characteristics such as soybean flowering time, disease resistance and stress resistance, and accurately analyzing these structural variations was of great significance for improving soybean yield, quality and other agronomic traits.

Professor Dajian ZHANG is guiding the students' experiments
Prof. Dajian ZHANG has been working on identification of the domestication genes of important agronomic traits in soybean and dissecting their molecular mechanisms for many years. By now, he has published many papers in the peer reviewed journals such as Nature Plants, Molecular Plant, Journal of Integrative Plant Biology and high impacted academic journals as the first or corresponding authors (including co-authors). Combined with the content of the previous research, in view of the important genes that have been discovered for quality and yield, the research team has bred high-yield and high-oil soybean varieties suitable for planting in the Huang-Huai-Hai region by using molecular marker assistant technology, and the new variety has participated in the regional test of soybeans in Shandong Province, performed well.
Yongbin ZHUANG, associate Prof. of SDAU, is the first author of this paper, and Dajian ZHANG, associate Prof. of SDAU, and Jianxin MA,PurdueUniversity,are the co-corresponding authors of this paper. Prof. Xiansheng ZHANG of SDAU and Prof. Jeffrey J. Doyle of Cornell University, Dr. Jacob B. Landis, Prof. Scott A. Jackson, University of Georgia, USA, Dr. Steven B. Cannon, USDA Corn Insect and Crop Genetics Institute, And Dr. Jane Grimwood, Alpha Biotechnology Institute, Hudson, USA, Dr. Jeremy Schmutz and Dr. Xutong WANG of Purdue University participated in the study. The research was co-funded by the National Key Research and Development Program of China, Shandong Province Improved Varieties Project, Taishan Scholars and Experts Program, and the Plant Genome Research Program of the National Science Foundation of the United States.

 Chinese
Chinese